MAIN OUTPUTS
Main output files
Output data structure
output_dir
├── 105923 # Subject ID
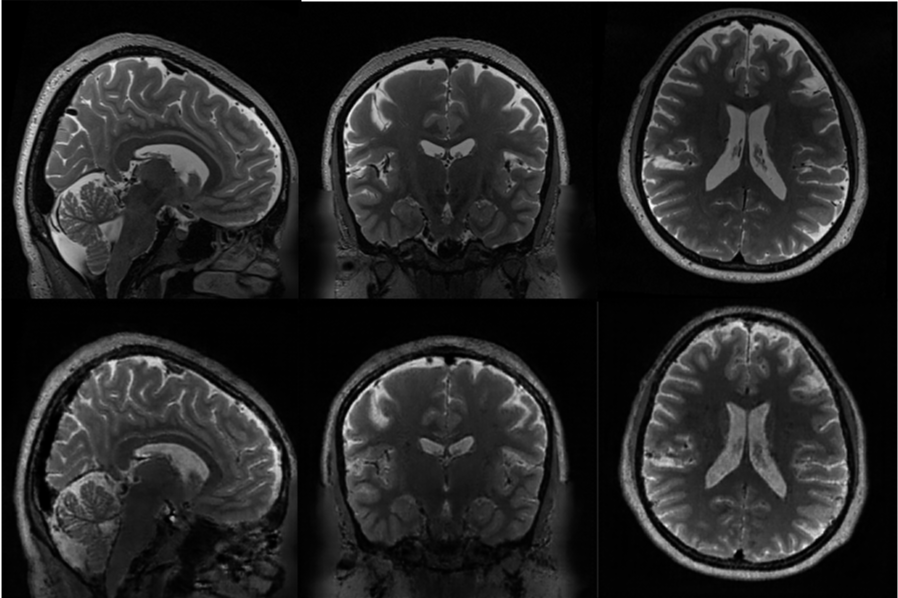
│ ├── T2w.nii.gz # Synthesized T2-weighted MRI
│ ├── myelin.nii.gz # Synthesized microstructure-sensitive proxy
│ ├── schaefer-100_MPC_matrix.txt # MPC matrix with schaefer-100 atlas
│ ├── schaefer-200_MPC_matrix.txt # MPC matrix with schaefer-200 atlas
│ ├── schaefer-300_MPC_matrix.txt # MPC matrix with schaefer-300 atlas
│ ├── schaefer-400_MPC_matrix.txt # MPC matrix with schaefer-400 atlas
│ ├── schaefer-500_MPC_matrix.txt # MPC matrix with schaefer-500 atlas
│ ├── schaefer-600_MPC_matrix.txt # MPC matrix with schaefer-600 atlas
│ ├── schaefer-700_MPC_matrix.txt # MPC matrix with schaefer-700 atlas
│ ├── schaefer-800_MPC_matrix.txt # MPC matrix with schaefer-800 atlas
│ ├── schaefer-900_MPC_matrix.txt # MPC matrix with schaefer-900 atlas
│ ├── schaefer-1000_MPC_matrix.txt # MPC matrix with schaefer-1000 atlas
│ ├── vosdewael-100_MPC_matrix.txt # MPC matrix with vosdewael-100 atlas
│ ├── vosdewael-200_MPC_matrix.txt # MPC matrix with vosdewael-200 atlas
│ ├── vosdewael-300_MPC_matrix.txt # MPC matrix with vosdewael-300 atlas
│ ├── vosdewael-400_MPC_matrix.txt # MPC matrix with vosdewael-400 atlas
│ ├── aparc_MPC_matrix.txt # MPC matrix with aparc atlas
│ ├── aparc-a2009s_MPC_matrix.txt # MPC matrix with aparc-a2009s atlas
│ ├── economo_MPC_matrix.txt # MPC matrix with economo atlas
│ ├── glasser-360_MPC_matrix.txt # MPC matrix with glasser-360 atlas
│ ├── schaefer-100_MPC_gradients.txt # Microstructural gradient with schaefer-100 atlas
│ ├── schaefer-200_MPC_gradients.txt # Microstructural gradient with schaefer-200 atlas
│ ├── schaefer-300_MPC_gradients.txt # Microstructural gradient with schaefer-300 atlas
│ ├── schaefer-400_MPC_gradients.txt # Microstructural gradient with schaefer-400 atlas
│ ├── schaefer-500_MPC_gradients.txt # Microstructural gradient with schaefer-500 atlas
│ ├── schaefer-600_MPC_gradients.txt # Microstructural gradient with schaefer-600 atlas
│ ├── schaefer-700_MPC_gradients.txt # Microstructural gradient with schaefer-700 atlas
│ ├── schaefer-800_MPC_gradients.txt # Microstructural gradient with schaefer-800 atlas
│ ├── schaefer-900_MPC_gradients.txt # Microstructural gradient with schaefer-900 atlas
│ ├── schaefer-1000_MPC_gradients.txt # Microstructural gradient with schaefer-1000 atlas
│ ├── vosdewael-100_MPC_gradients.txt # Microstructural gradient with vosdewael-100 atlas
│ ├── vosdewael-200_MPC_gradients.txt # Microstructural gradient with vosdewael-200 atlas
│ ├── vosdewael-300_MPC_gradients.txt # Microstructural gradient with vosdewael-300 atlas
│ ├── vosdewael-400_MPC_gradients.txt # Microstructural gradient with vosdewael-400 atlas
│ ├── aparc_MPC_gradients.txt # Microstructural gradient with aparc atlas
│ ├── aparc-a2009s_MPC_gradients.txt # Microstructural gradient with aparc-a2009s atlas
│ ├── economo_MPC_gradients.txt # Microstructural gradient with economo atlas
│ ├── glasser-360_MPC_gradients.txt # Microstructural gradient with glasser-360 atlas
│ ├── schaefer-100_MPC_moment.txt # Microstructural moment with schaefer-100 atlas
│ ├── schaefer-200_MPC_moment.txt # Microstructural momnet with schaefer-200 atlas
│ ├── schaefer-300_MPC_moment.txt # Microstructural momnet with schaefer-300 atlas
│ ├── schaefer-400_MPC_moment.txt # Microstructural momnet with schaefer-400 atlas
│ ├── schaefer-500_MPC_moment.txt # Microstructural momnet with schaefer-500 atlas
│ ├── schaefer-600_MPC_moment.txt # Microstructural momnet with schaefer-600 atlas
│ ├── schaefer-700_MPC_moment.txt # Microstructural momnet with schaefer-700 atlas
│ ├── schaefer-800_MPC_moment.txt # Microstructural momnet with schaefer-800 atlas
│ ├── schaefer-900_MPC_moment.txt # Microstructural momnet with schaefer-900 atlas
│ ├── schaefer-1000_MPC_moment.txt # Microstructural momnet with schaefer-1000 atlas
│ ├── vosdewael-100_MPC_moment.txt # Microstructural momnet with vosdewael-100 atlas
│ ├── vosdewael-200_MPC_moment.txt # Microstructural momnet with vosdewael-200 atlas
│ ├── vosdewael-300_MPC_moment.txt # Microstructural momnet with vosdewael-300 atlas
│ ├── vosdewael-400_MPC_moment.txt # Microstructural momnet with vosdewael-400 atlas
│ ├── aparc_MPC_moment.txt # Microstructural momnet with aparc atlas
│ ├── aparc-a2009s_MPC_moment.txt # Microstructural momnet with aparc-a2009s atlas
│ ├── economo_MPC_moment.txt # Microstructural momnet with economo atlas
│ └── glasser-360_MPC_moment.txt # Microstructural momnet with glasser-360 atlas
└── ...
Synthesized T2-weighted MRI
Actual T2w (top) / Synthesized T2w (bottom)
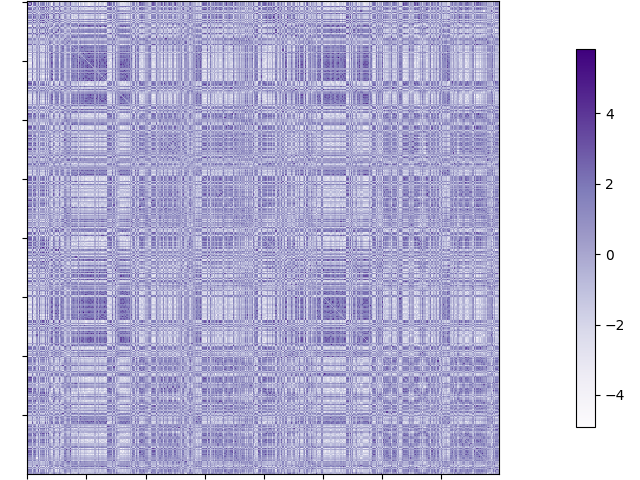
Synthesized MPC matrix
import numpy as np
from nilearn import plotting
# load matrix
matrix = np.loadtxt("~/output_dir/sub/schaefer-400_MPC_matrix.txt")
print(matrix.shape)
>>> (400, 400)
MPC matrix visualization.
plotting.plot_matrix(matrix, cmap='Purples')
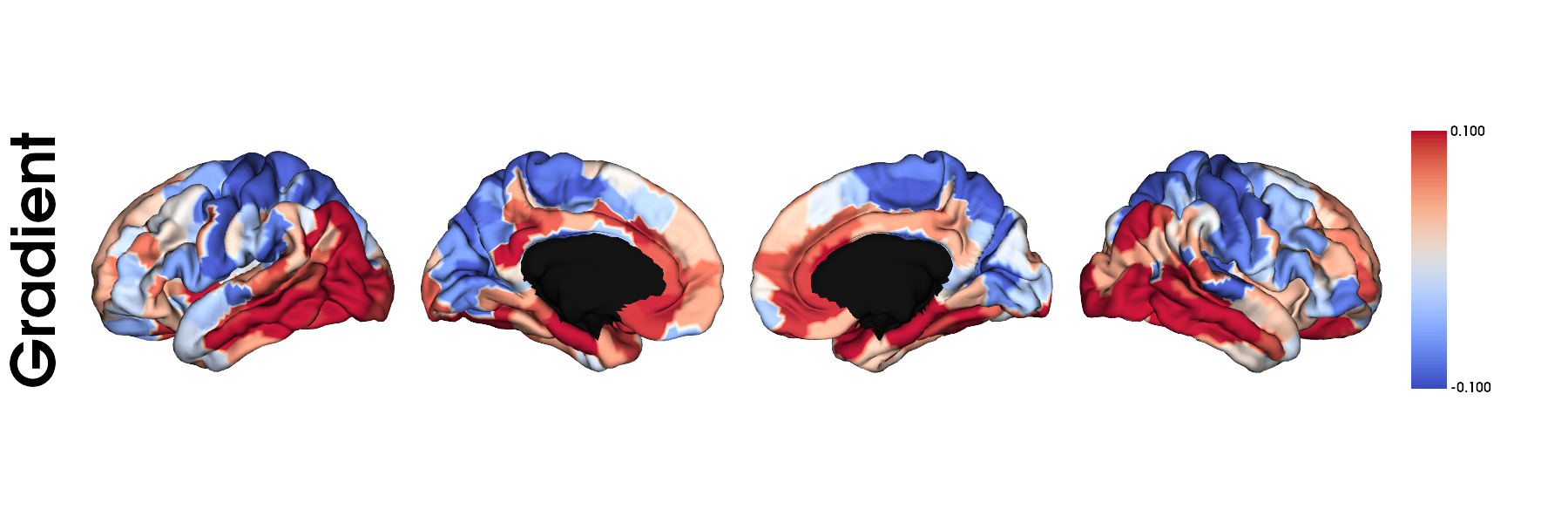
Synthesized microstructural gradient
We calculated gradients using diffusion embedding and normalized angle kernel with 0.9 sparsity. More details in /GAN-MAT/functions/preprocessing.py
import numpy as np
grads_400 = np.loadtxt("~/output_dir/sub/schaefer-400_MPC_gradients.txt")
print(grads_400.shape)
>>> (400, 10)
import nibabel as nib
from brainspace.datasets import load_fsa5
from brainspace.plotting import plot_hemispheres
from brainspace.utils.parcellation import map_to_labels
# Load the fsaverage5 surfaces
fs5_lh, fs5_rh = load_fsa5()
# Load annotation file in fsaverage5
annot_lh_fs5= nib.freesurfer.read_annot("~/GAN-MAT/parcellations/lh.schaefer-400_mics.annot")
annot_rh_fs5= nib.freesurfer.read_annot("~/GAN-MAT/parcellations/rh.schaefer-400_mics.annot")[0]+200
annot_rh_fs5 = np.where(annot_rh_fs5==200, 0, annot_rh_fs5)
labels_fs5 = np.concatenate((annot_lh_fs5[0], annot_rh_fs5), axis=0)
# Mask of the medial wall on fsaverage5
mask_fs5 = labels_fs5 != 0
# Map gradients to original parcels
grad = [None]
for i in range(1):
grad[i] = map_to_labels(-grads_400[:,i], labels_fs5, mask=mask_fs5, fill=np.nan)
# Plot gradients
plot_hemispheres(fs5_lh, fs5_rh, array_name=grad, size=(1800, 600), cmap='coolwarm',
color_bar=True, label_text=['Gradient'], zoom=1.2)
Synthesized microstructural moments
We provide microstructural moment features(mean, standard deviation, skewness, kurtosis).
moments_400 = np.loadtxt("~/output_dir/sub/schaefer-400_MPC_moment.txt")
print(moments_400.shape)
>>> (4, 400)
# Map moments to original parcels
moments = [None]
for i in range(4):
moments[i] = map_to_labels(moments_400[:,i], labels_fs5, mask=mask_fs5, fill=np.nan)
# Plot gradients
plot_hemispheres(fs5_lh, fs5_rh, array_name=moments, size=(1600, 1200), cmap='coolwarm',
color_bar=True, label_text=['Mean', "SD", "Skewness", "Kurtosis"], zoom=1.35)
